1.3. Example: Solve van der Pol equation using Python¶
In this example, we use Open Interfaces from Python to solve Van der Pol equation:
with parameter \(\mu = 1000\), on time interval \(t \in [0, 3000]\). Note that such a high value of \(\mu\) makes the system stiff, in a sense that it exhibits both fast and slow dynamics and explicit methods for solving initial-value problems are not efficient for such systems as they require very small time steps to maintain stability.
To solve this second-order ordinary differential equation, we first rewrite it as a system of two first-order ordinary differential equations:
where \(u_1 = x\) and \(u_2 = \frac{\mathrm d x}{\mathrm d t}\).
We implement the right-hand side of this system in the class VdPEquationProblem
to avoid passing parameter \(\mu\) to the function that computes the right-hand
side:
class VdPEquationProblem:
def __init__(self, mu=5.0):
self.mu = mu
def compute_rhs(self, __, u, udot, ___):
udot[0] = u[1]
udot[1] = self.mu * (1 - u[0] ** 2) * u[1] - u[0]
return 0
Note that the __init__ method of this class accepts the parameter \(\mu\),
that is then used in the compute_rhs method.
Also note that the compute_rhs method has a fixed signature
defined by the IVP interface, therefore, it return 0 at the end
to indicate successful computation, which is not “pythonic”, indeed,
but is used for interoperability with other languages.
Having defined the right-hand side of the system, we can set other parameters of the initial-value problem: namely, initial condition and time span:
u0 = [2.0, 0.0] # Initial condition
t0 = 0 # Initial time
tfinal = 3000 # Final time
problem = VdPEquationProblem(mu=1000)
After that we load an implementation of the IVP interface. For example, we can use the implementation based on SciPy library:
solver = IVP("scipy_ode")
and pass the problem definition to the solver as long as required tolerances:
solver.set_initial_value(u0, t0)
solver.set_rhs_fn(problem.compute_rhs)
solver.set_tolerances(rtol=1e-8, atol=1e-12)
Let’s suppose that we require the solution at 501 equally spaced
points in the time interval [t0, tfinal]:
times = np.linspace(problem.t0, tfinal, num=501)
Finally, we try to integrate the system and store the solution:
solution = np.empty((len(times), len(y0)))
solution[0] = y0
i = 1
for t in times[1:]:
solver.integrate(t)
solution[i] = s.y
i += 1
If we run the code, we receive the following error message:
/home/user/sw/miniforge3/envs/open-interfaces/lib/python3.13/site-packages/scipy/integrate/_ode.py:438: UserWarning: dopri5: larger nsteps is needed
self._y, self.t = mth(self.f, self.jac or (lambda: None),
Traceback (most recent call last):
File "/home/dima/Work/um02-open-interfaces/lang_python/oif_impl/openinterfaces/_impl/ivp/scipy_ode/scipy_ode.py", line 62, in integrate
assert self.s.successful()
~~~~~~~~~~~~~~~~~^^
AssertionError
[bridge_python] Call failed
[dispatch] ERROR: During execution of the function 'ivp::integrate' an error occurred
due to the fact that the default maximum number of steps is not allowed to solve this stiff problem.
Even if we increase the maximum number of steps allowed:
solver.set_integrator("dopri5", {"nsteps": 100_000})
and integrate again, we receive the same error message.
Even, if we switch to another solver available in SciPy,
for example, vode with bdf method:
solver.set_integrator("vode", {"method": "bdf"})
we still receive the same error message due to the stiffness of the problem.
We have to increase the maximum number of steps allowed for the vode solver
to a high value, for example, 40 000 steps:
solver.set_integrator("vode", {"method": "bdf", "nsteps": 40_000})
and only then, the solver is able to solve the problem and we arrive at the solution Fig. 1.3.1, although integration takes a while.
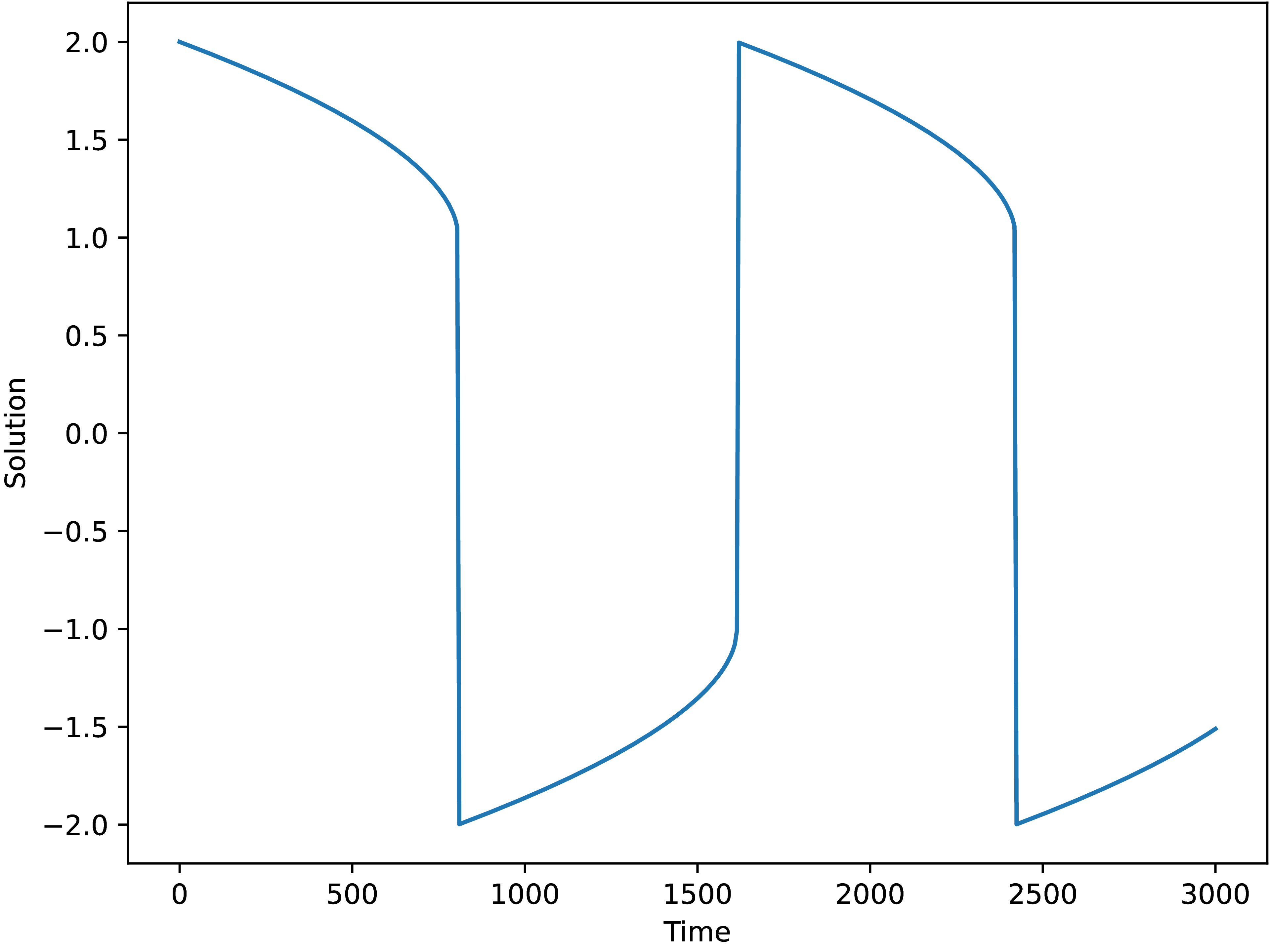
Fig. 1.3.1 Solution of the Van der Pol equation with \(mu=1000\)
using scipy_ode with the vode integrator.¶
Finally, we try to use other implementations of the IVP interface,
for example, Rosenbrok23 integrator from the OrdinaryDiffEq.jl Julia
package, which in Open Interfaces is available via the jl_diffeq
implementation:
solver = IVP("jl_diffeq")
solver.set_integrator("Rosenbrock23", {"autodiff": False,})
(here we replace Automatic Differentiation with Forward Differencing
by supplying an integration option autodiff=False;
we are working currently on enabling use of Automatic Differentiation
in jl_diffeq). Running the code with this implementation,
we arrive at the solution quickly, see Fig. 1.3.2.
One can see that the solution is the same as the one obtained
with the vode solver from SciPy, Fig. 1.3.1.
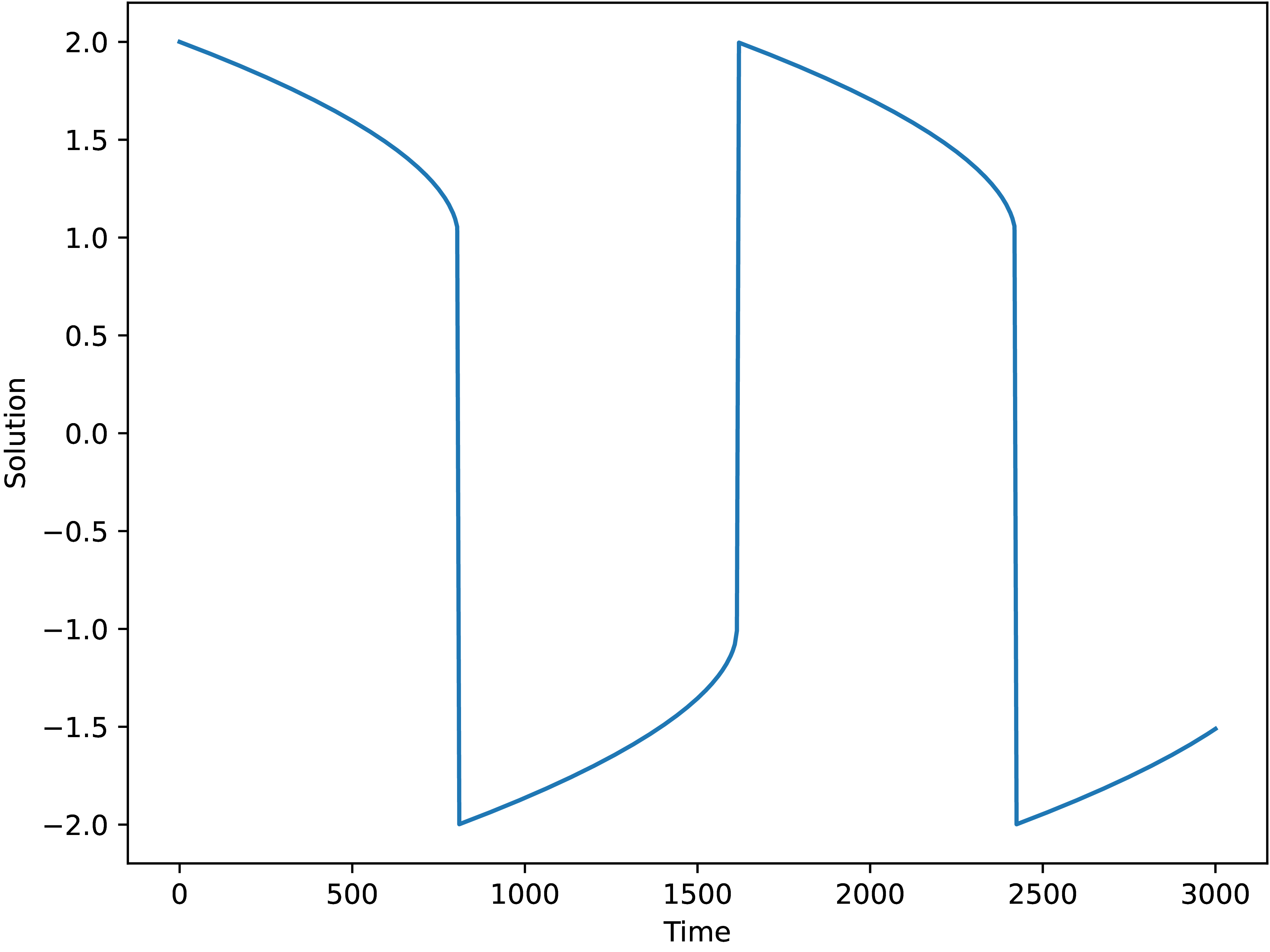
Fig. 1.3.2 Solution of the Van der Pol equation with mu=1000
using jl_diffeq implementation with the Rosenbrok23 integrator.¶
The full code of this example is available in the
examples/call_qeq_from_python.py and can be run as follows:
python examples/call_qeq_from_python.py [implementation]
where the implementation argument is one of the following:
scipy_ode-dopri5,scipy_ode-dopri5-100k,scipy_ode-vode,scipy_ode-vode-40k,sundials_cvode,jl_diffeq-rosenbrock23.
At the end of the computations, if they are successful, the resultant solution is displayed.